Leaf-colonising Bacteria Colony Phenotype
May 22, 2020 22:59 · 400 words · 2 minute read
Here are some bacteria I am working with. I will try to update some interesting info about these bacteria.
These bacteria were subcultured on R2A agar.
Phylogenetic tree
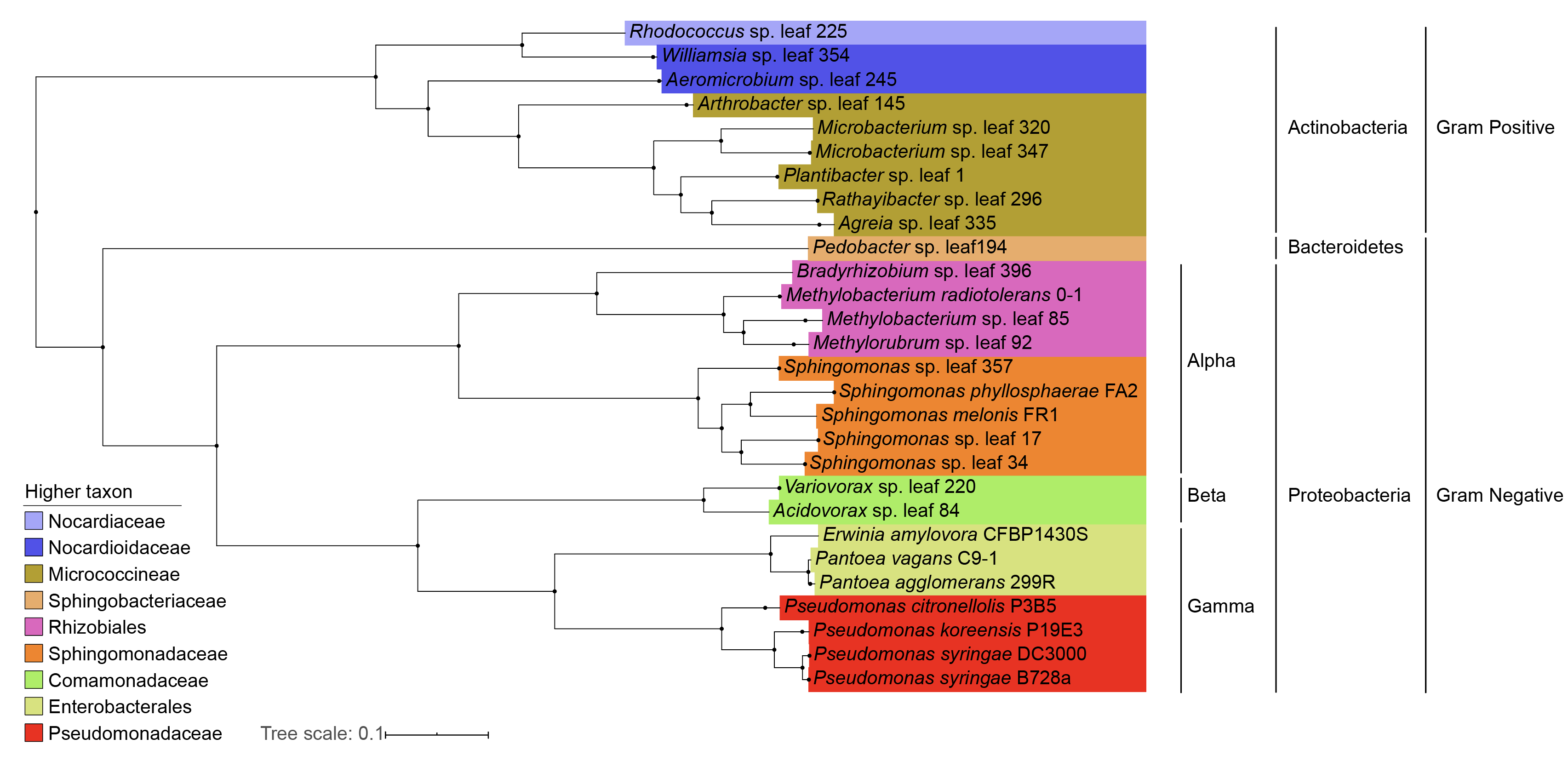 Retrived from my MSc thesis
Retrived from my MSc thesis
Here is a picture showing brief pigment and colony phenotype.
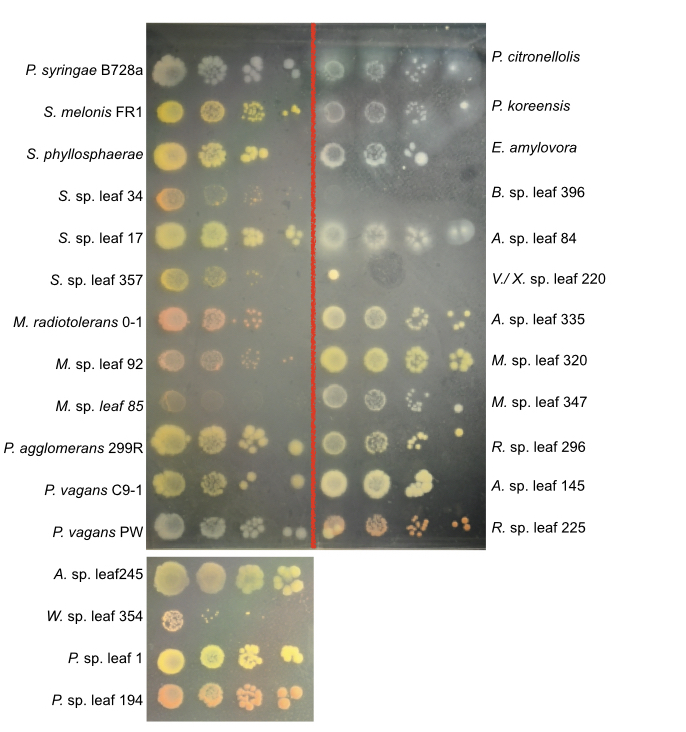 Full names will be listed below respectively.
Full names will be listed below respectively.
Genus Sphingomonas
Sphingomonas phyllosphaerae FA2
Sphingomonas melonis Fr1
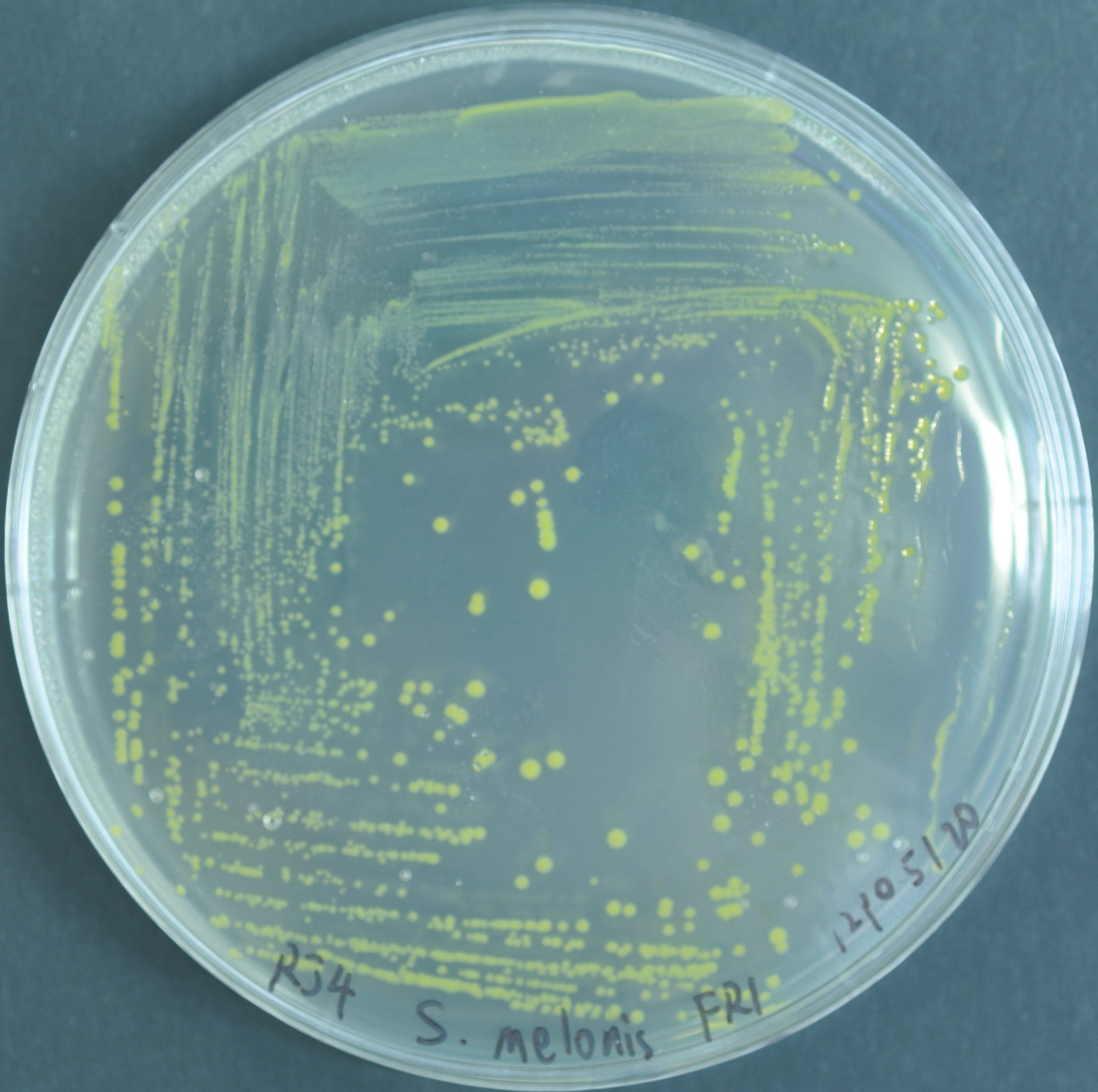
Due to the effect of mScarlt fluorescence, the pigment of bacteria colonies is also changed.
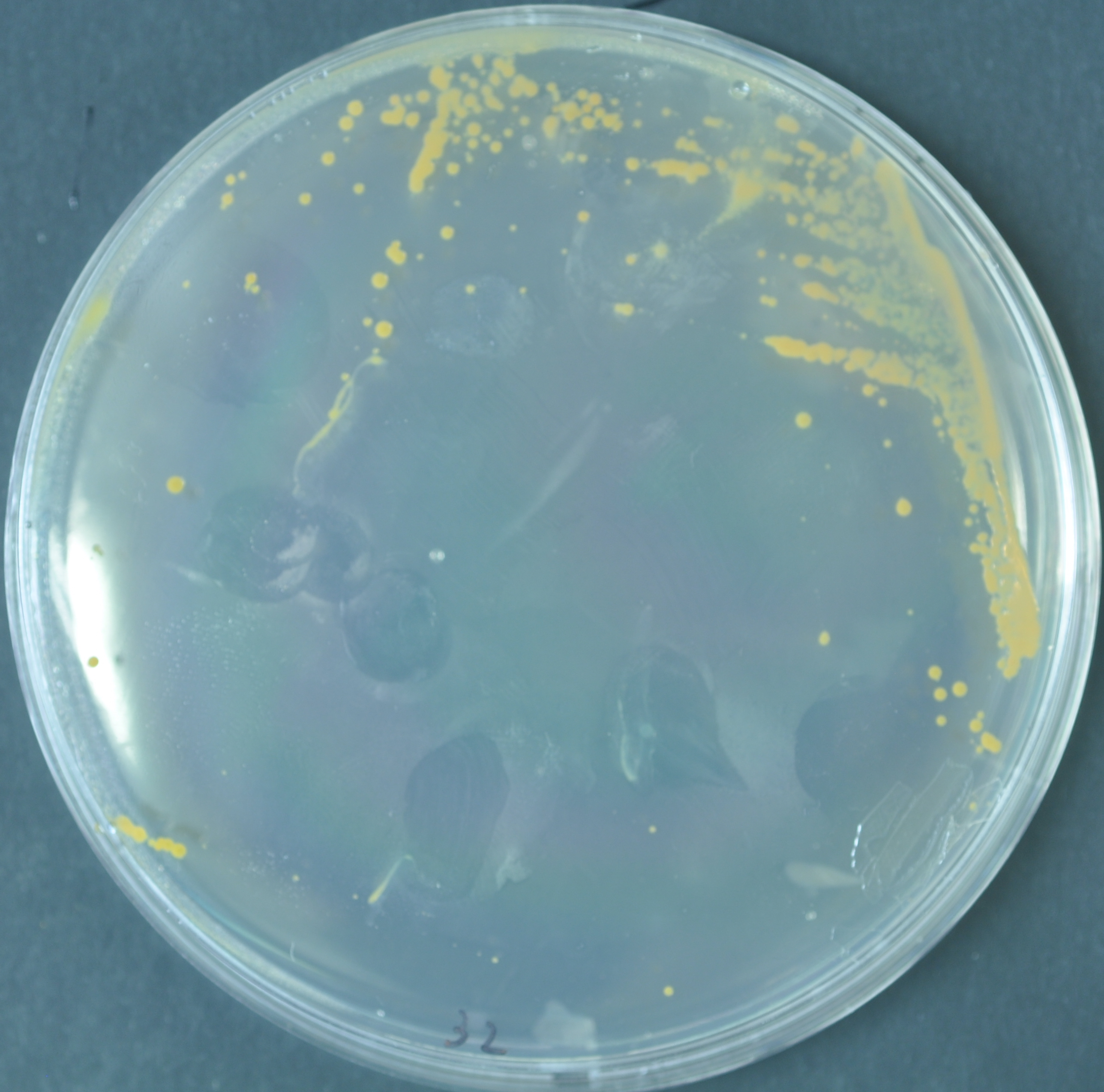
Sphingomonas sp. leaf 34
Sphingomonas sp. leaf 17
Sphingomonas sp. leaf 357
Genus Methylobacterium
Methylobacterium sp. leaf 85
Methylobacterium radiotolerans sp. 0-1
Methylobacterium sp. leaf 92

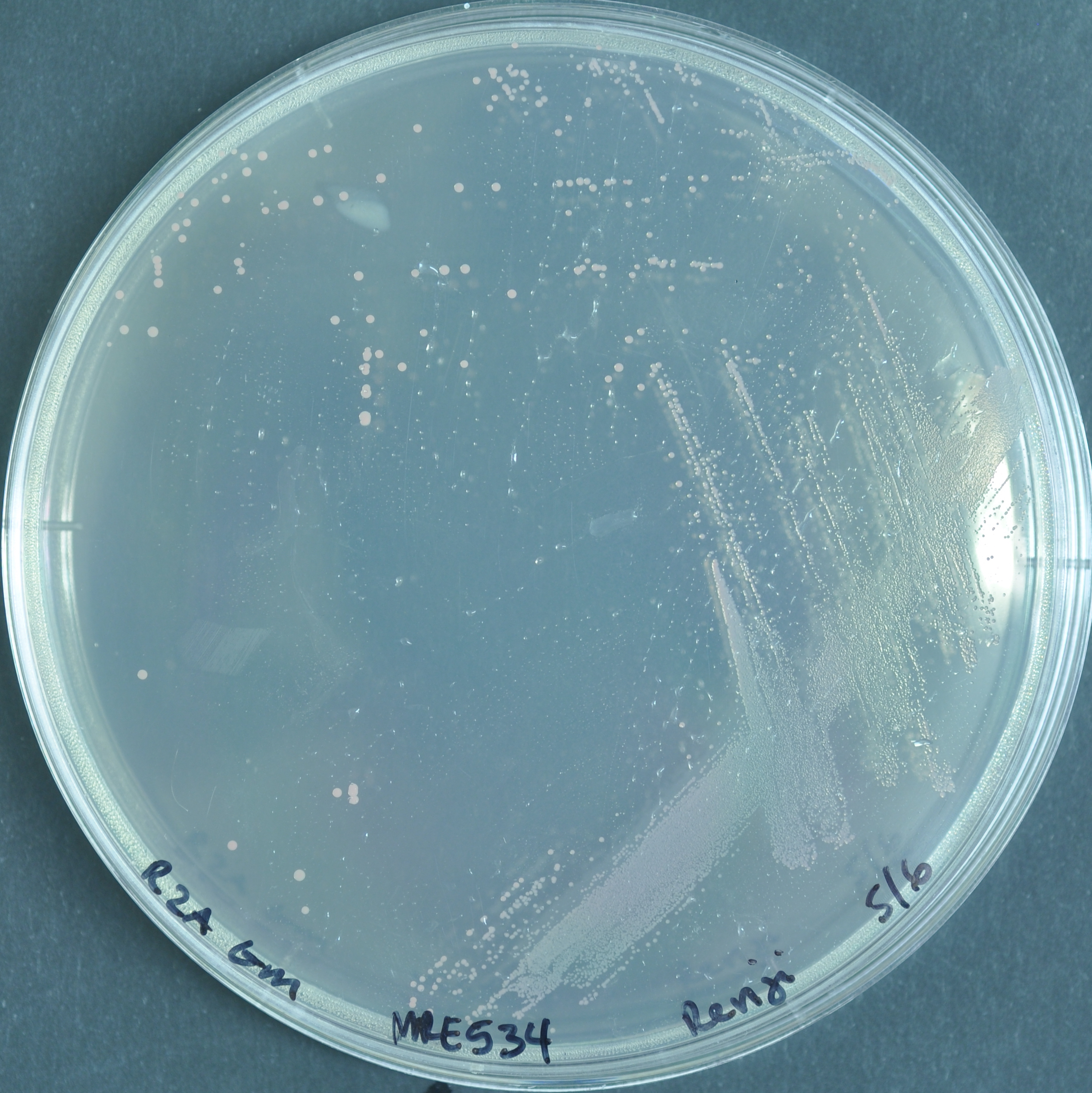
Acidovorax sp. leaf 84
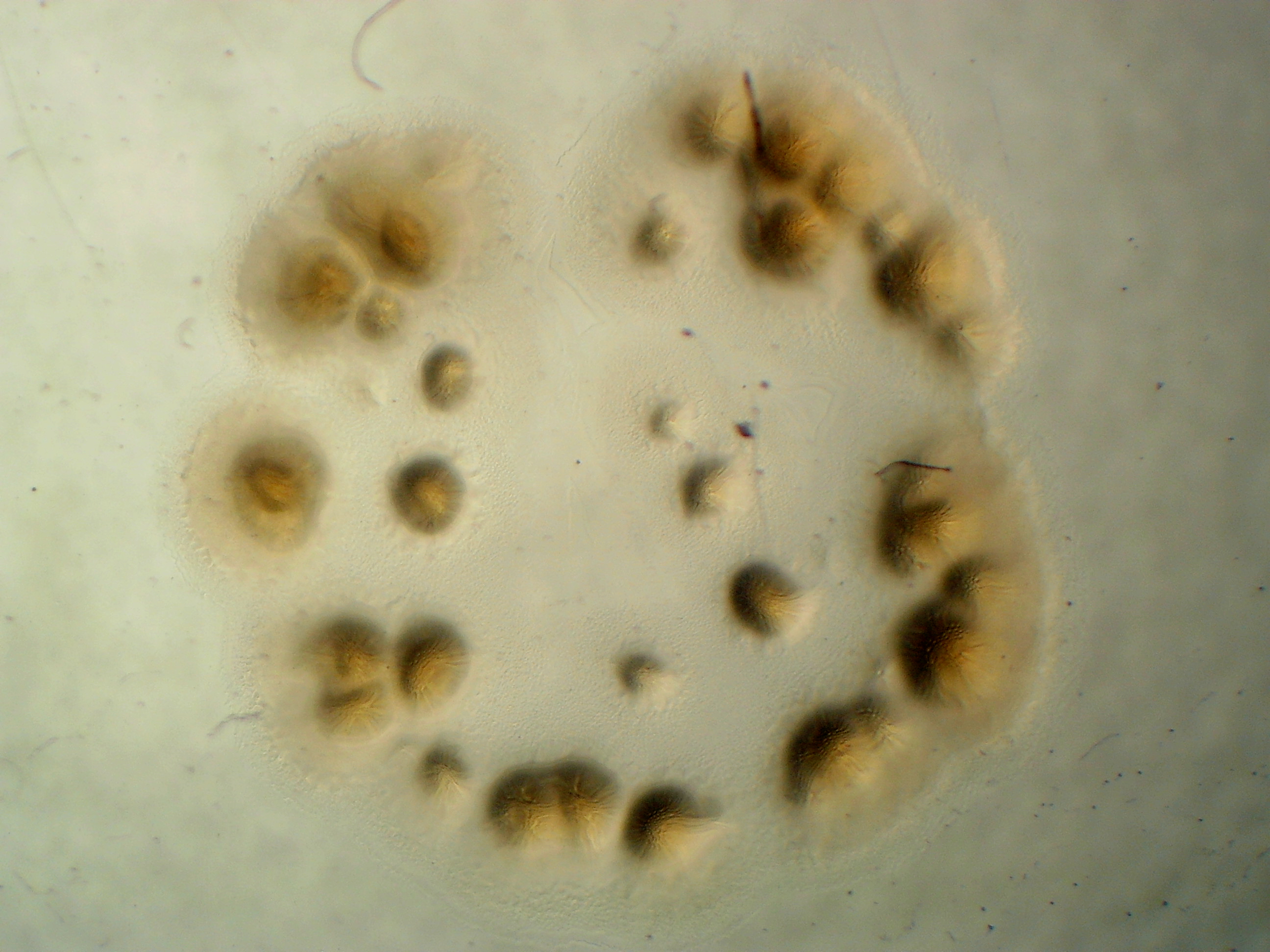
The pigment for fluorescent A. sp. leaf 84 does not change.
Genus Pseudomonas
Pseudomonas syringae B728a
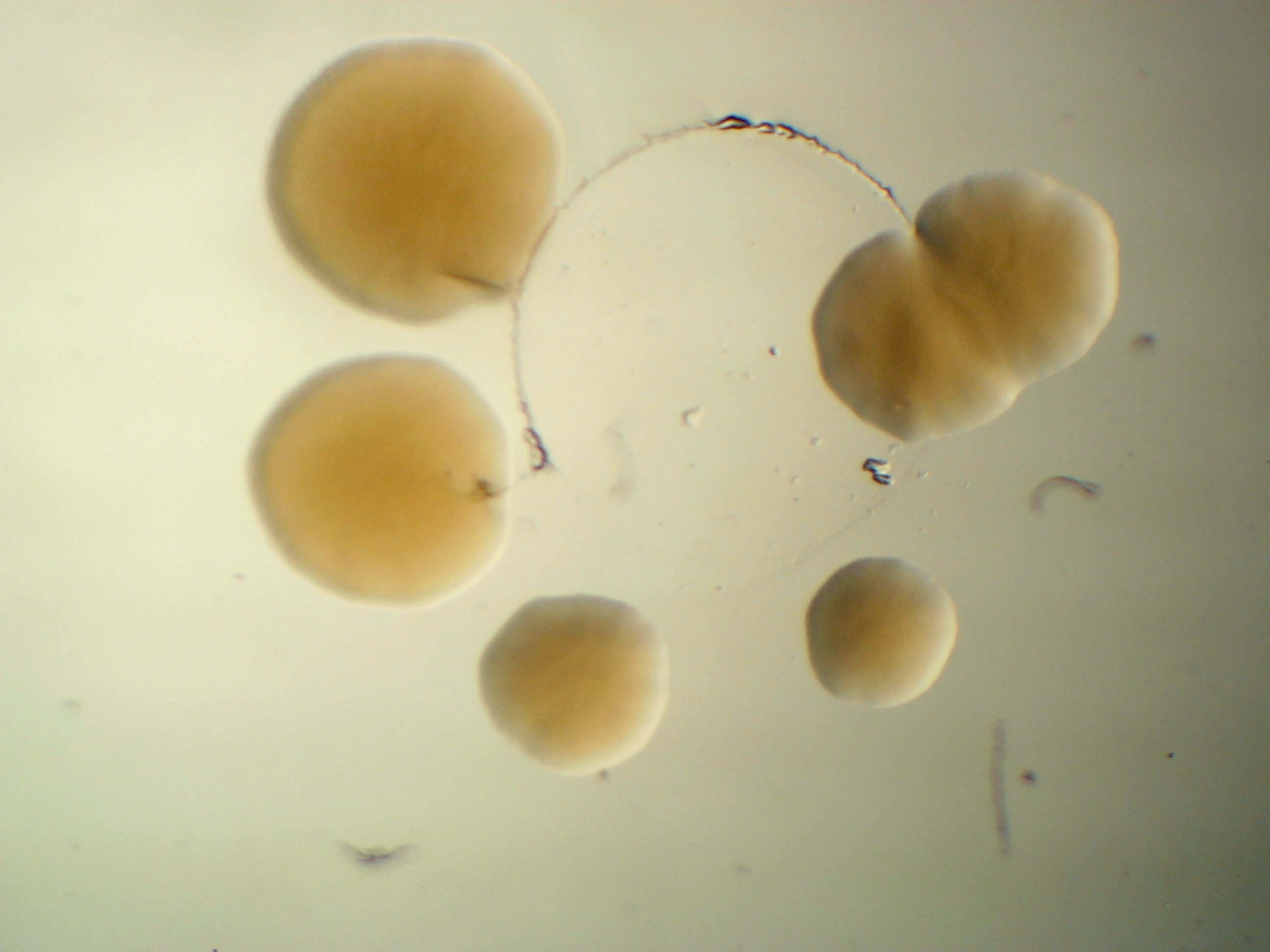
Pseudomonas syringae B728a is a bean pathogen and it was claimed that it is non-pathogenic to Arabidopsis thaliana (Vinatzer et al. 2006). However, in my experiment, it does show a negative effect on Arabidopsis growth.
Pseudomonas citronellolis
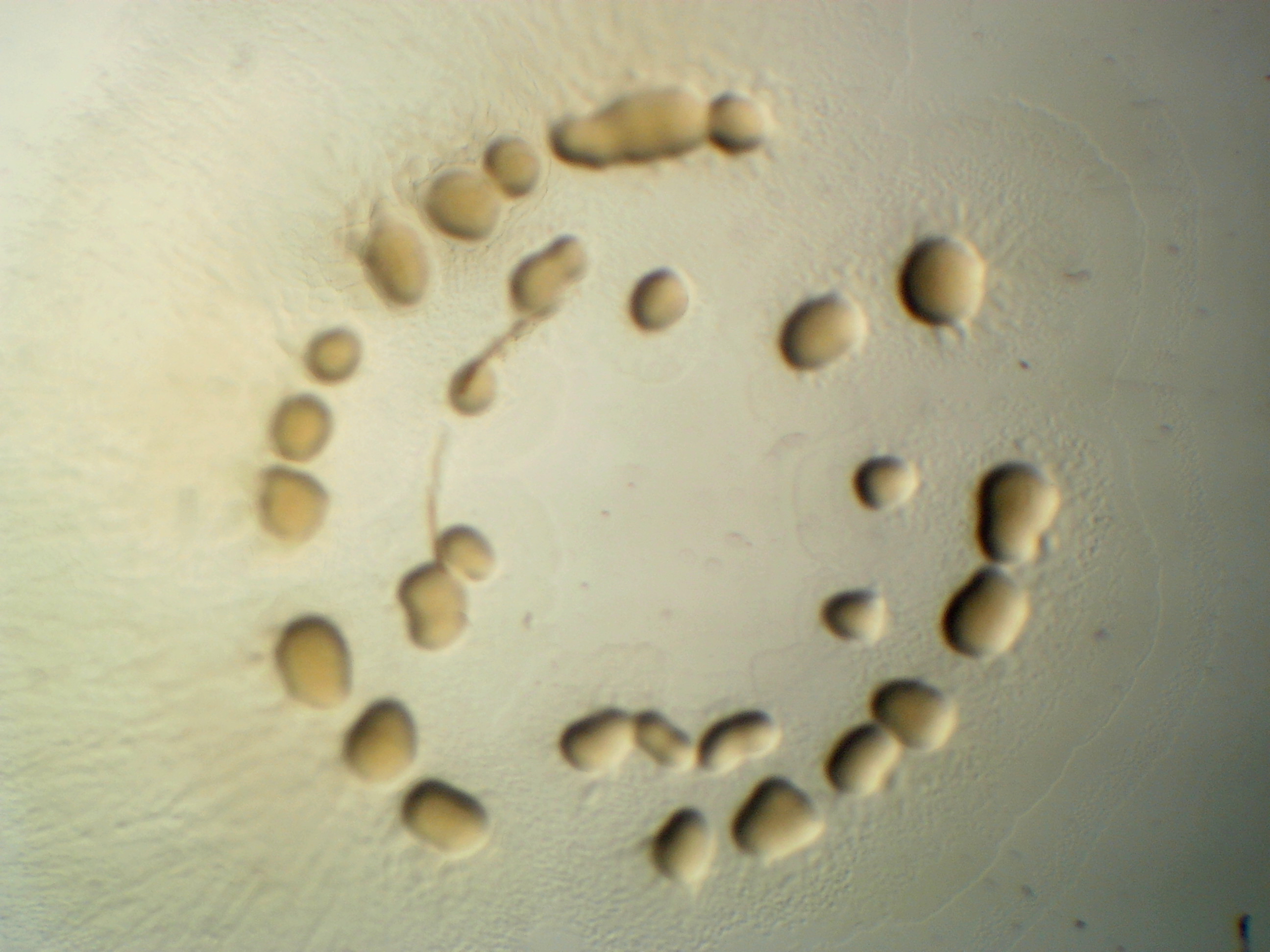
For P. cironellolis, now it is little pinkish.
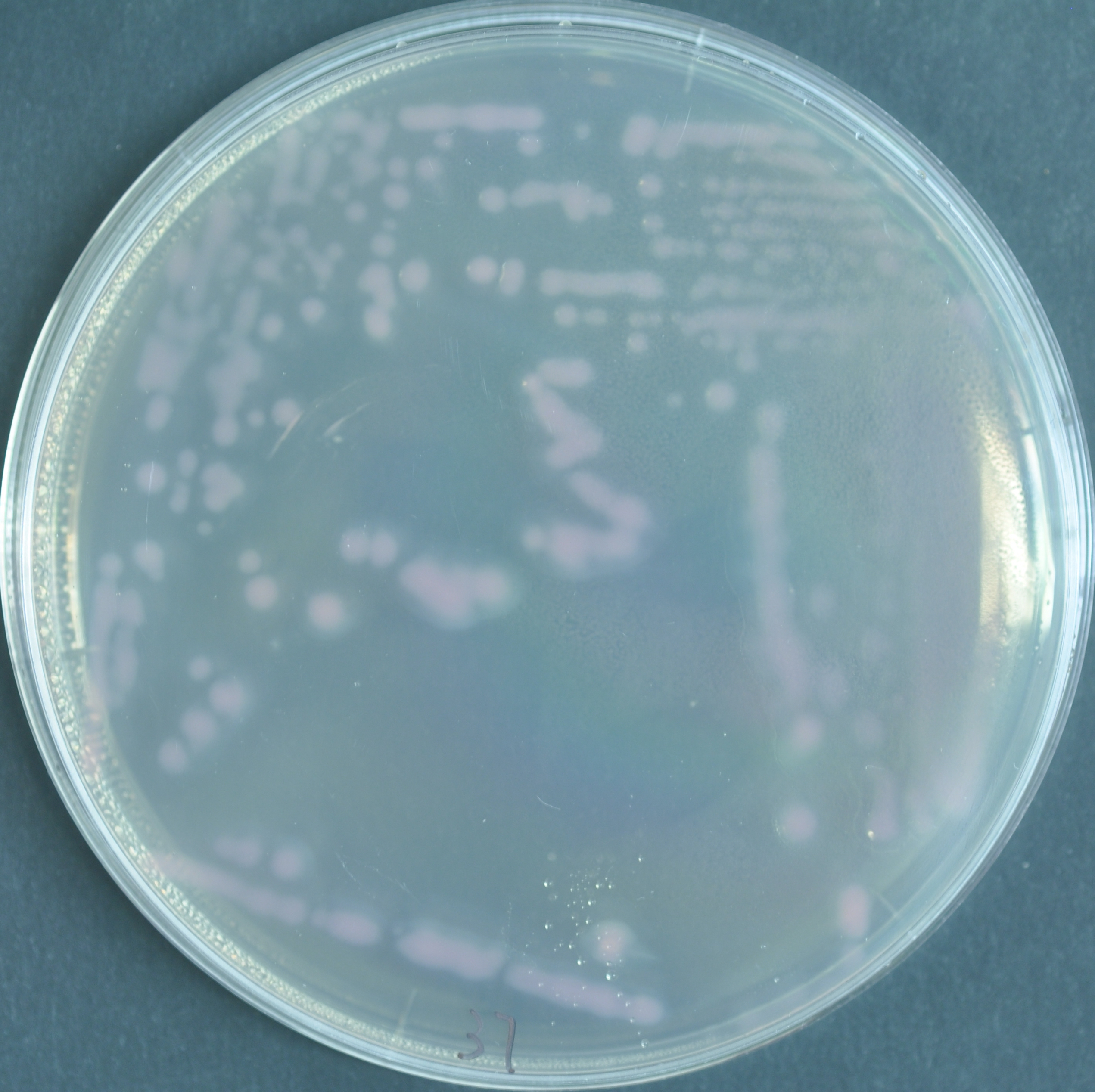
Pseudomonas koreensis

Pantoea agglomerans 299R
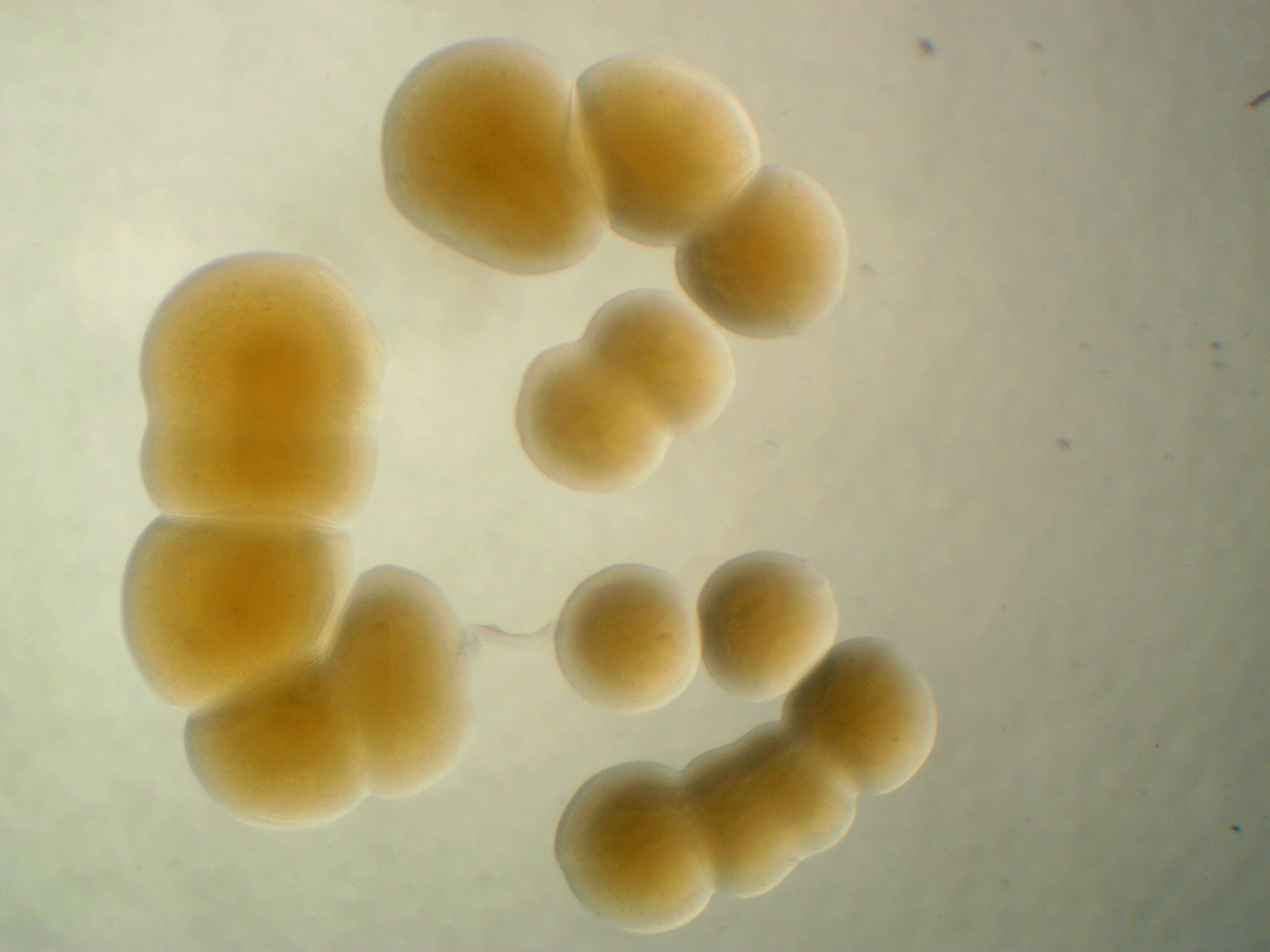
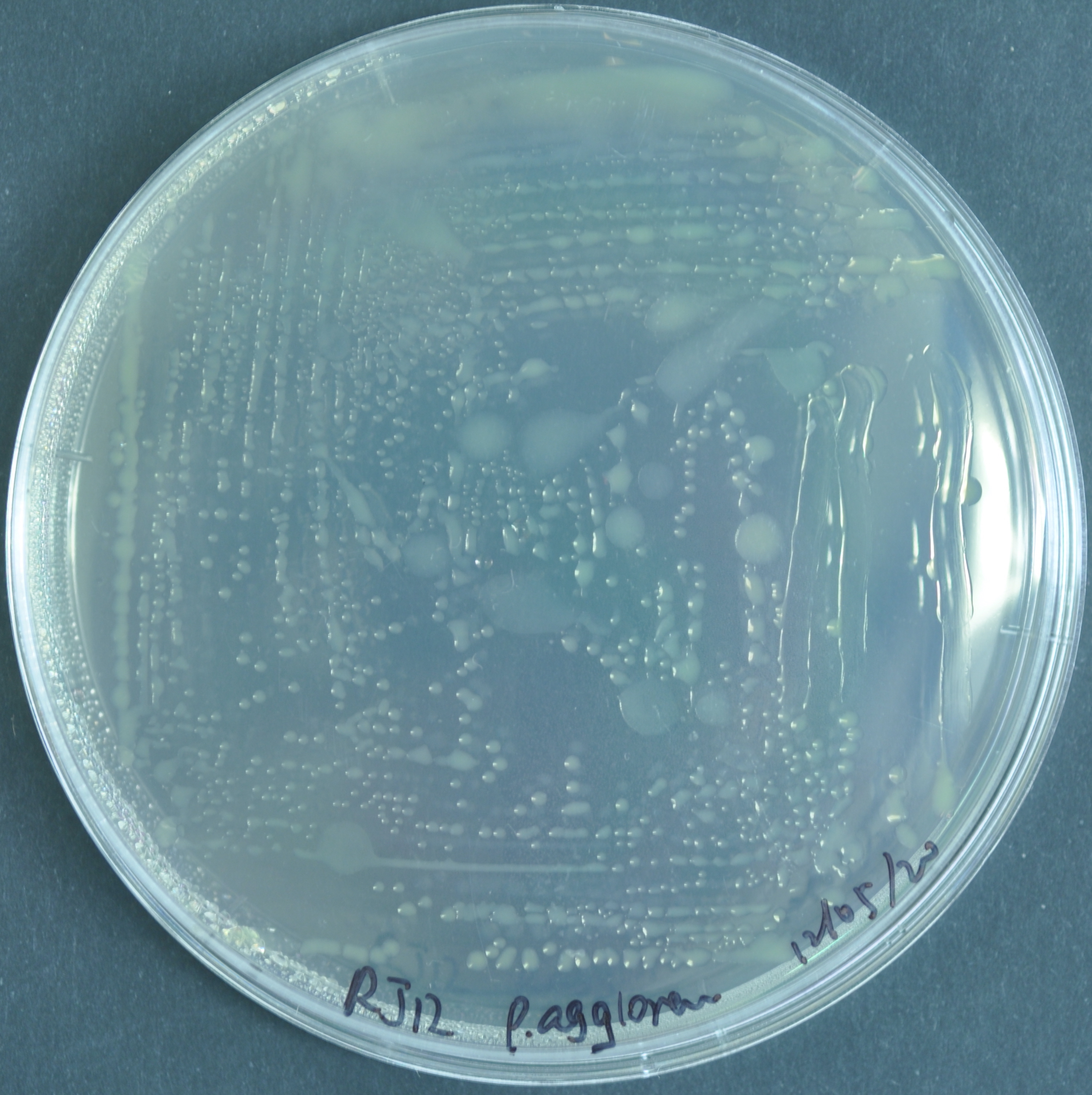
However, the pigment turns orange for genus Pantoea.
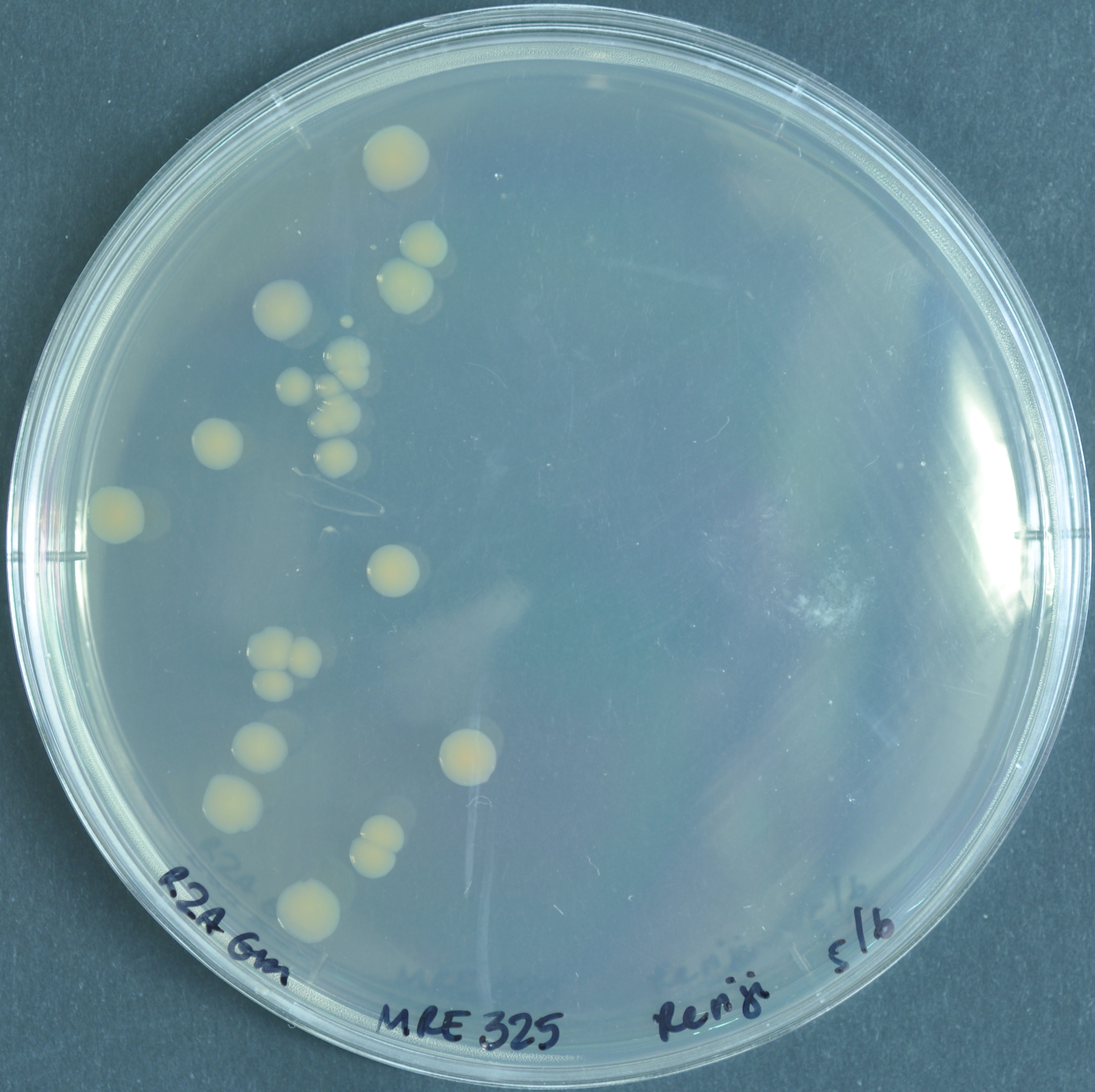
Pantoea vegans C9-1
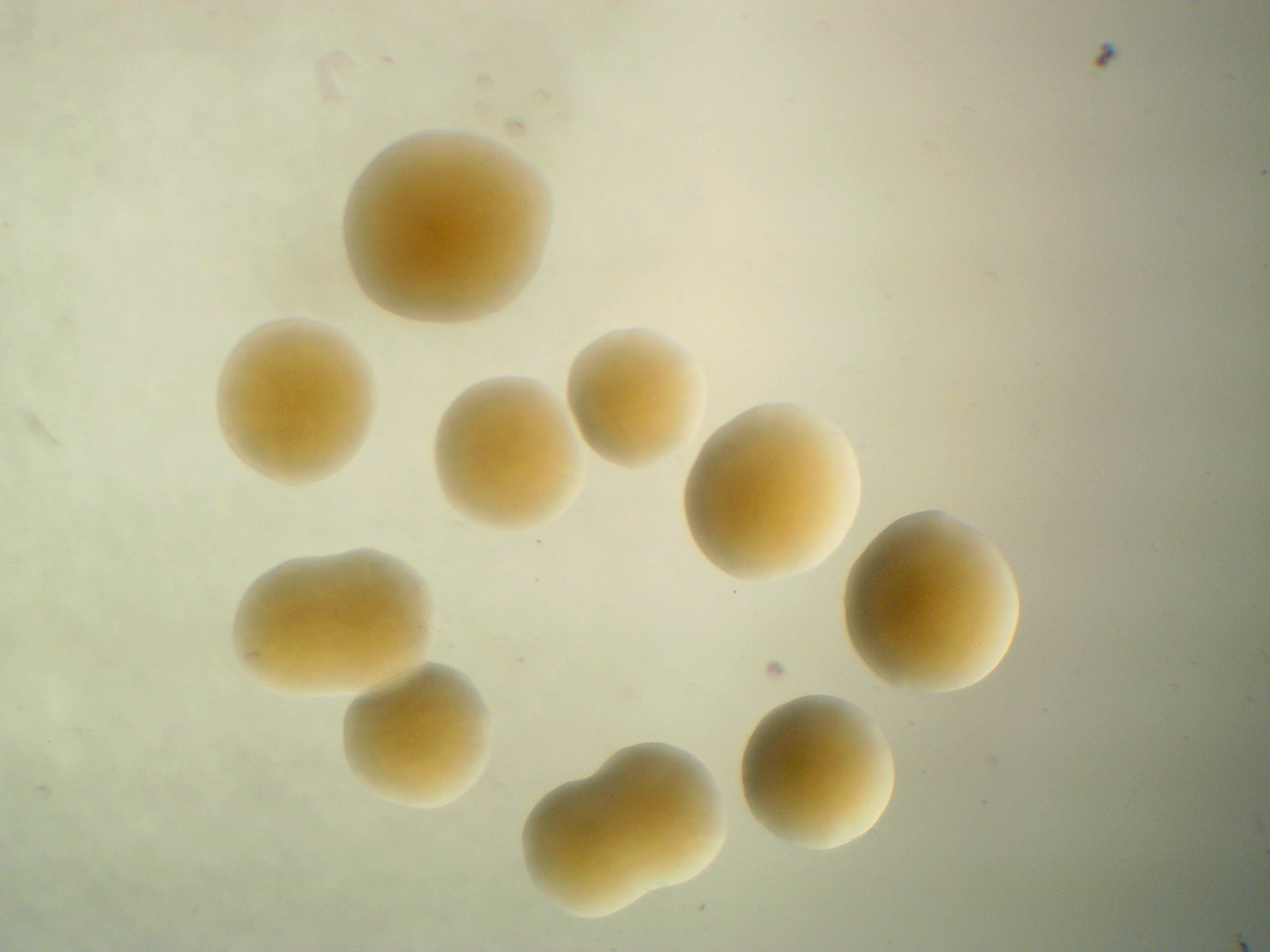
Pantoea vegans PW
 Pantoea vagans PW is a white variant of C9-1 and misses a mega plasmid.
Pantoea vagans PW is a white variant of C9-1 and misses a mega plasmid.
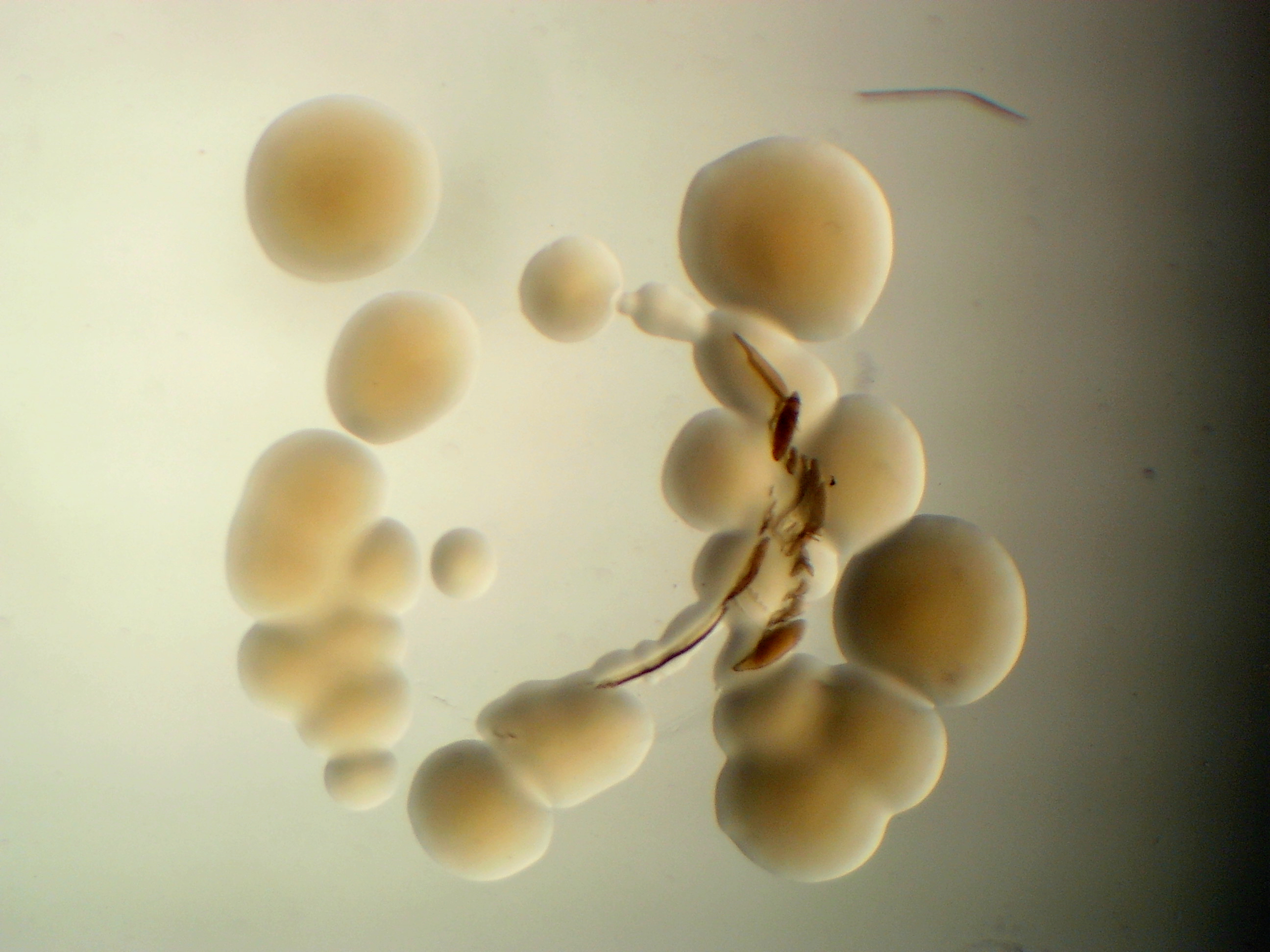
Williamsia sp. leaf 354
Rhodococcus leaf 225
Aeromicrobium sp. leaf 245
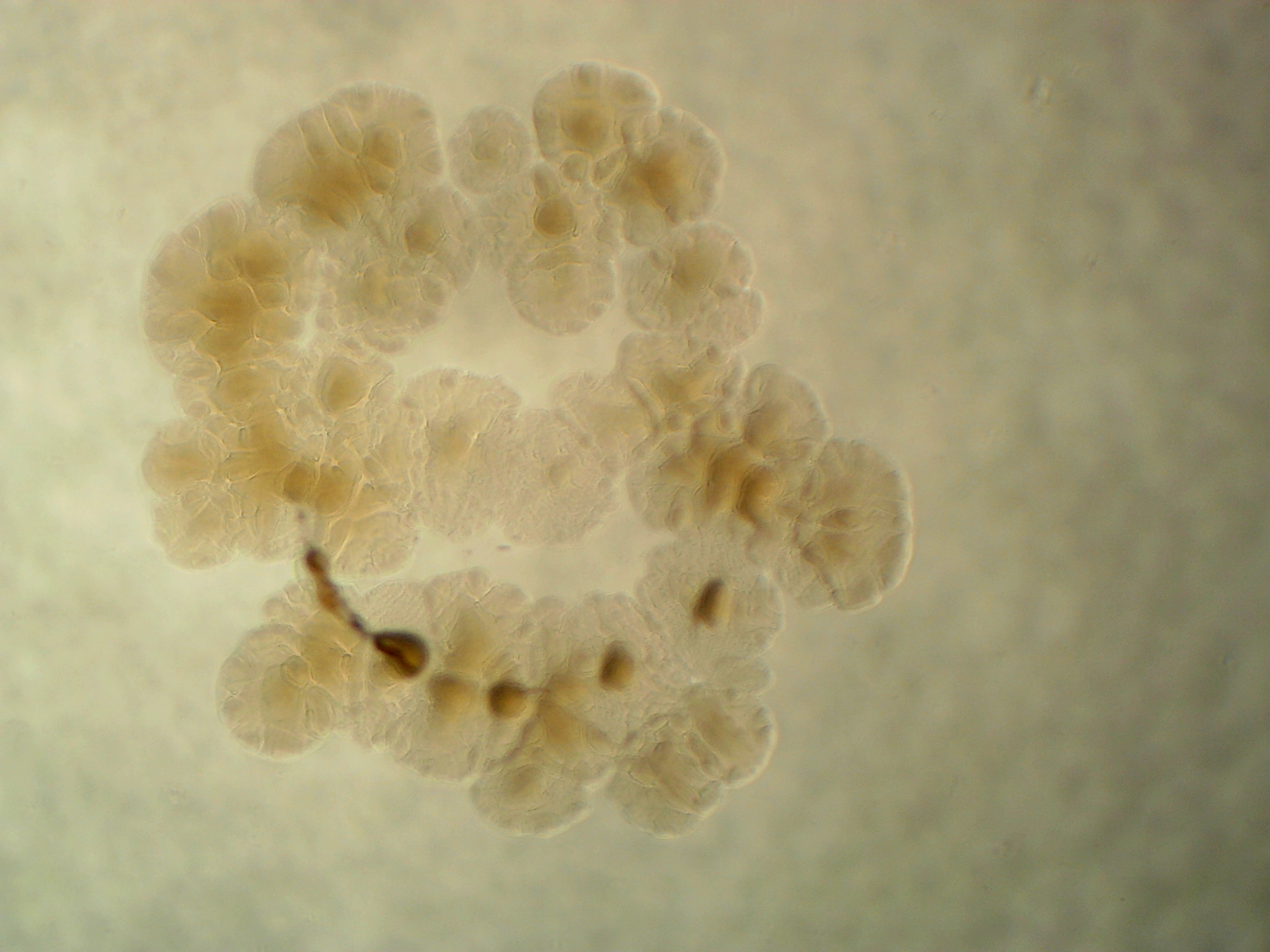
Arthrobacter sp. leaf 145
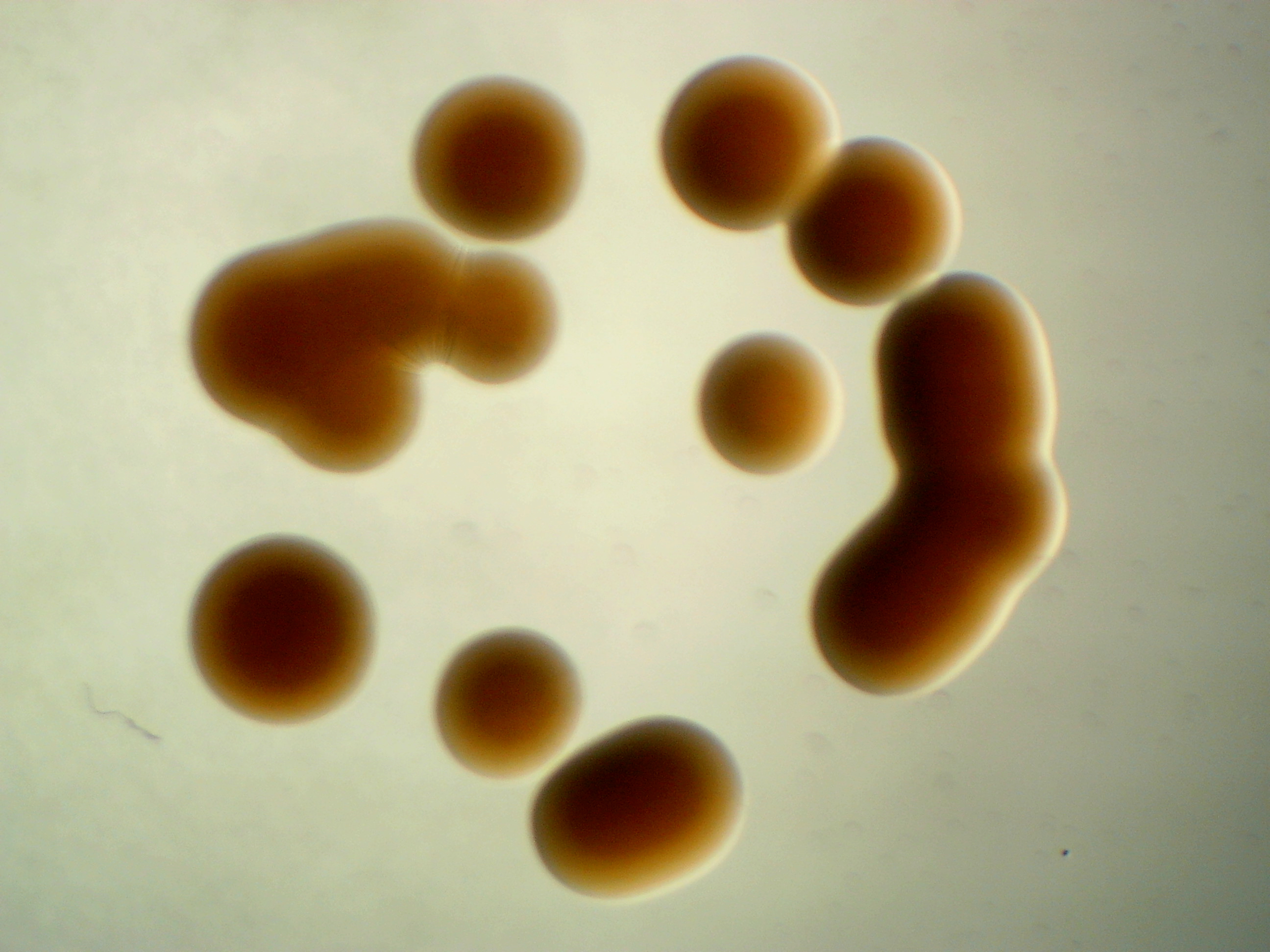
Microbacterium sp. leaf 320
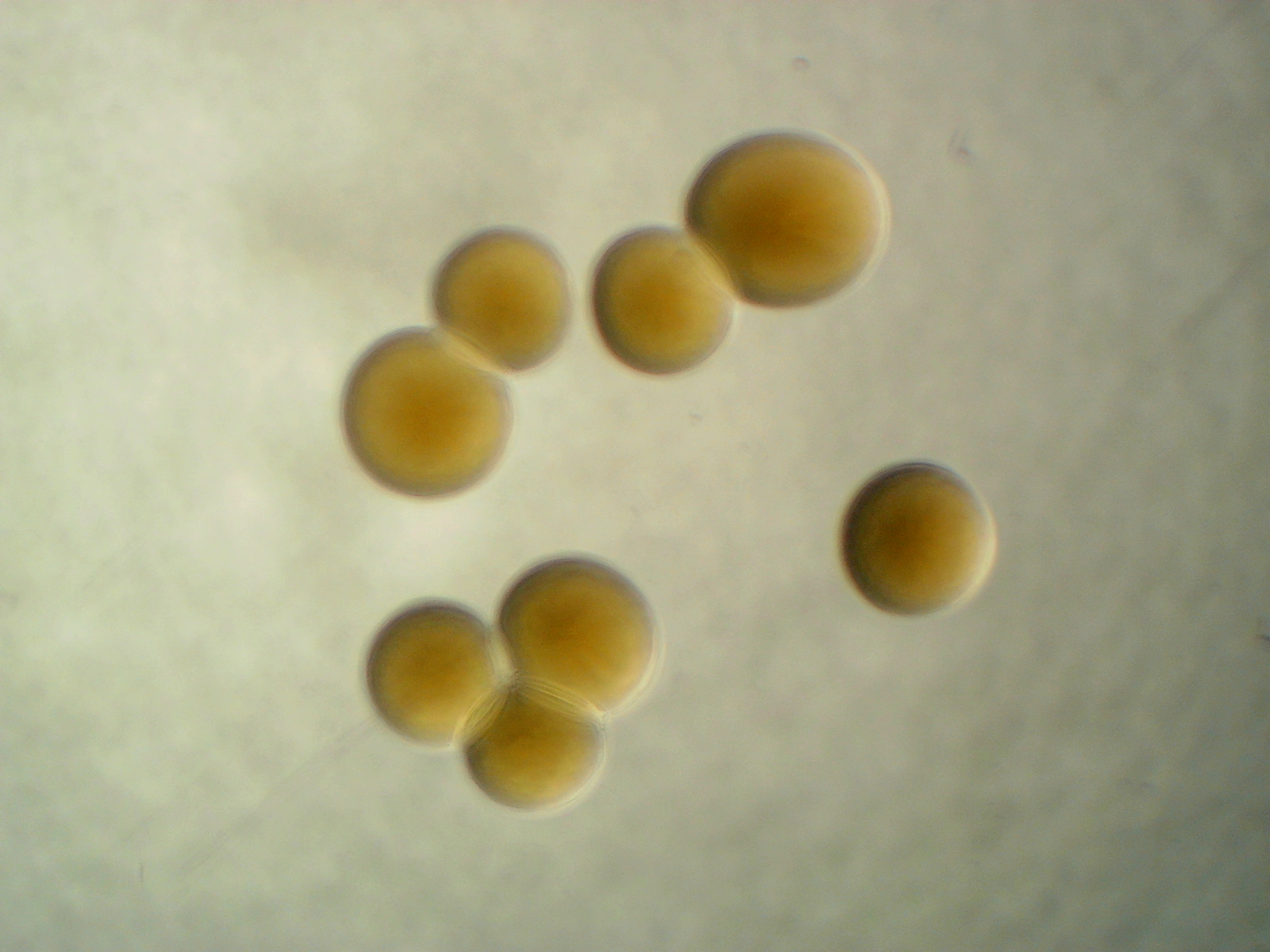
Microbacterium sp. leaf 347
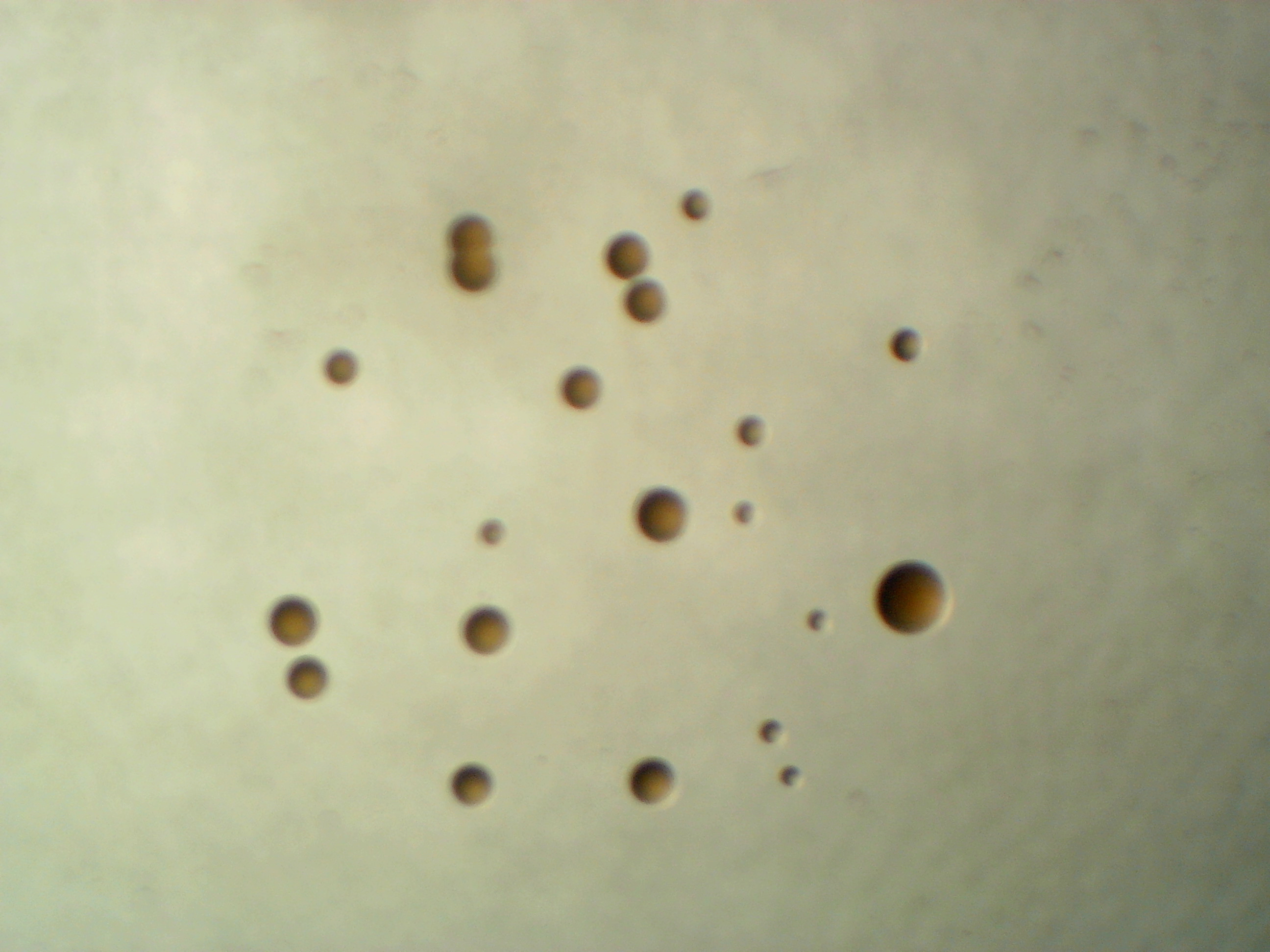
Plantibacter sp. leaf 1
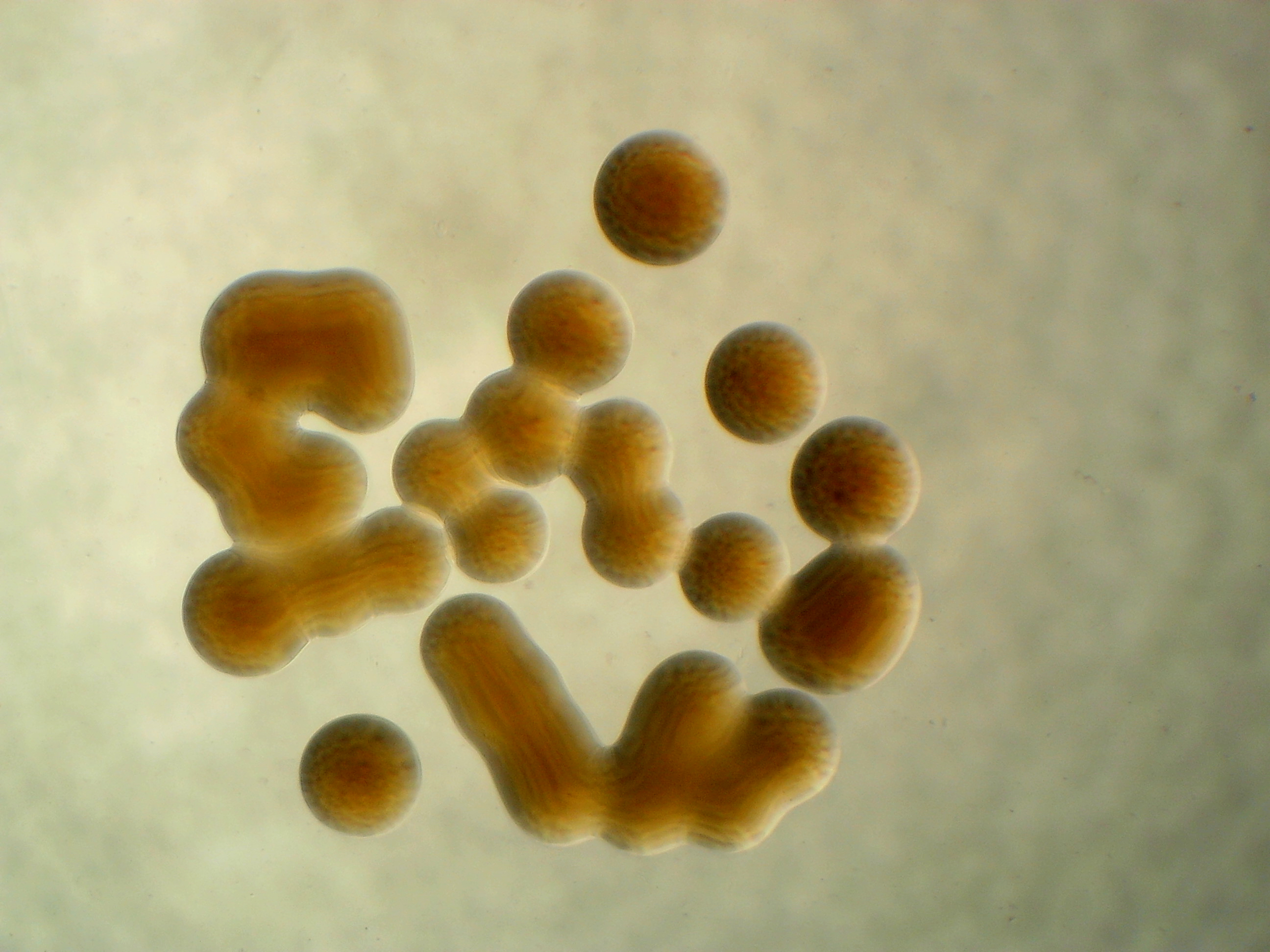
Rathayibacter sp. leaf 296
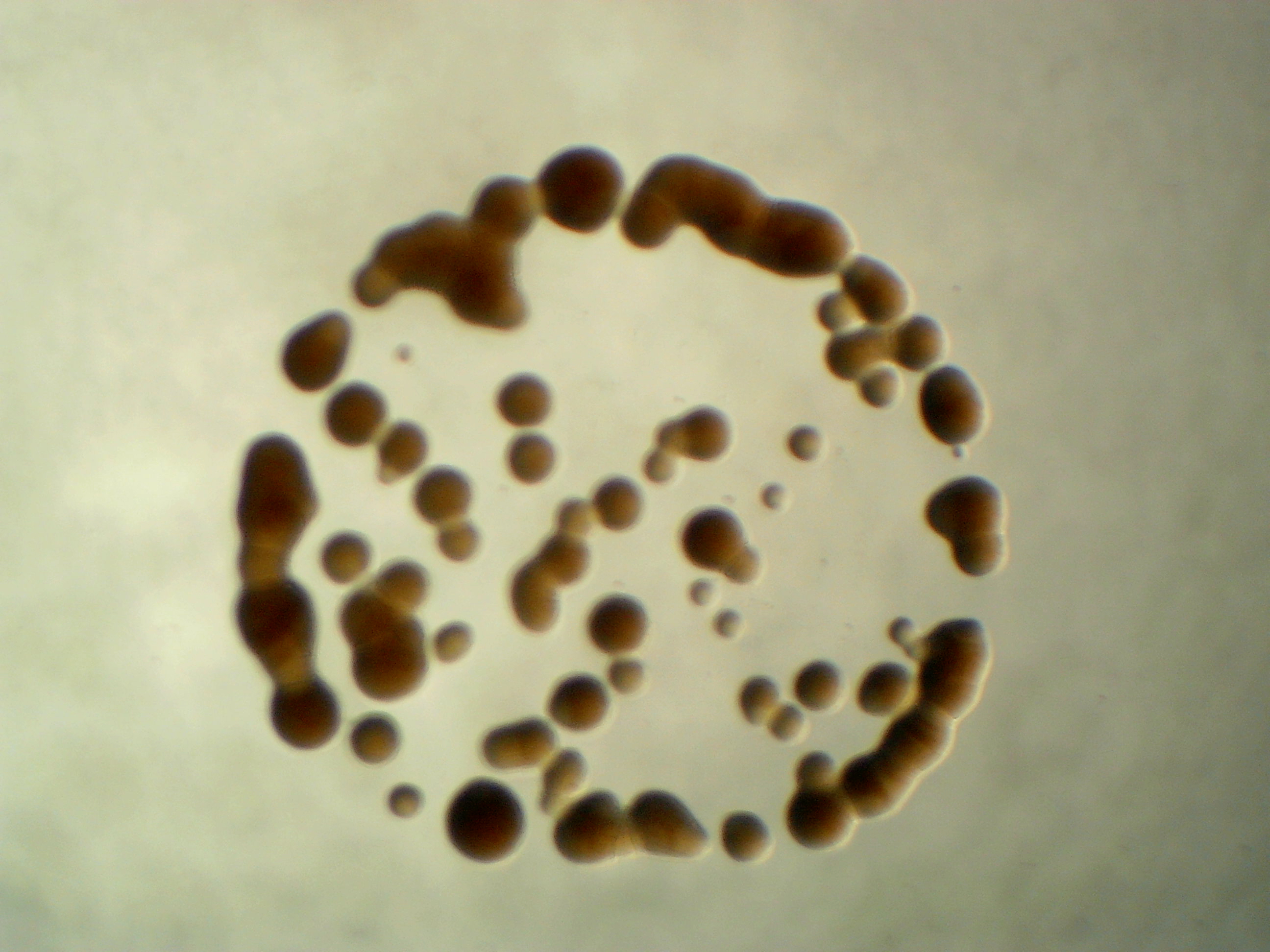
Agreia sp. leaf 335
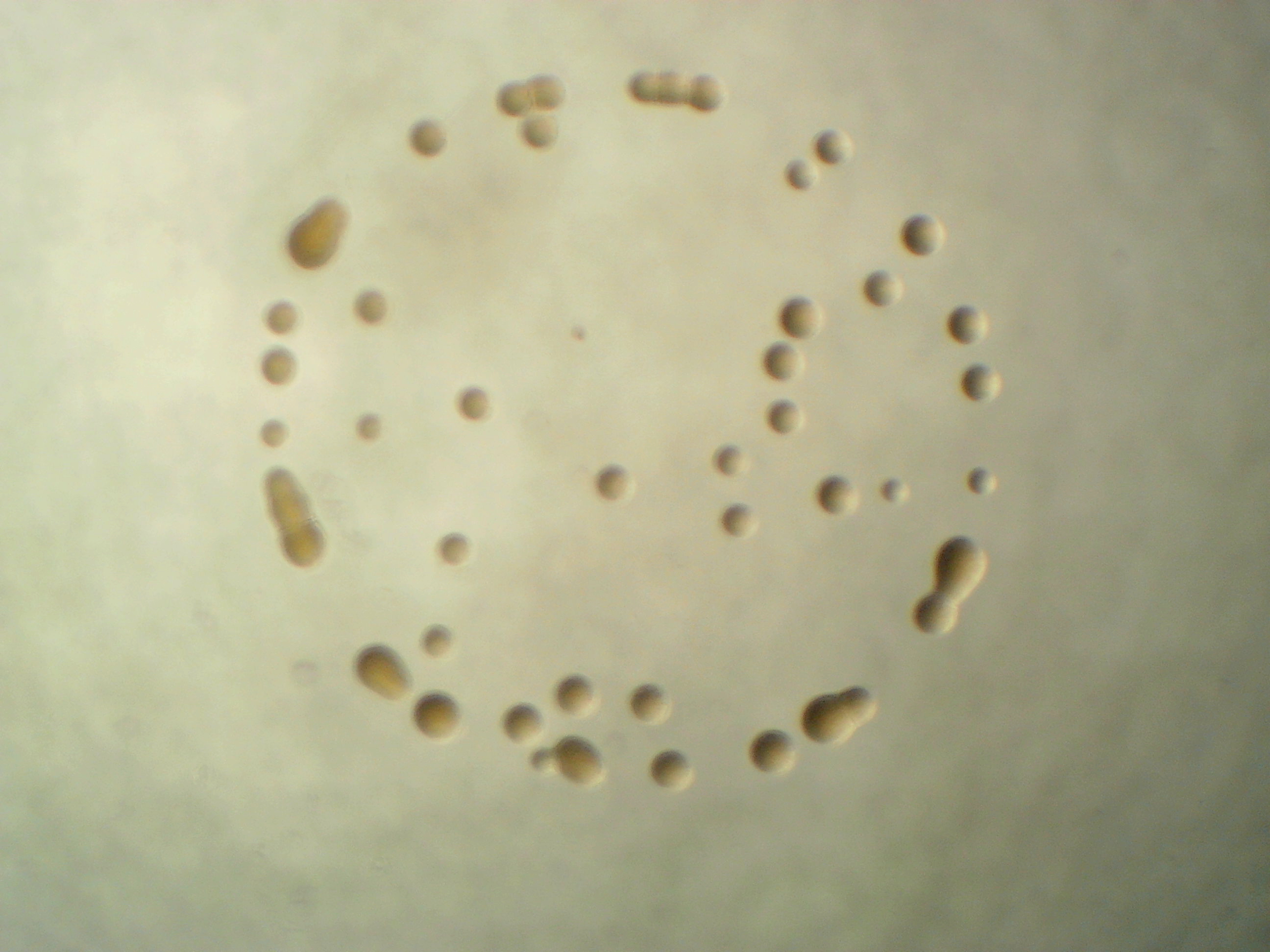
Erwinia amylovora

Bradyrhizobium sp. leaf 396
Pedobacter sp. leaf 194

Xylophilus sp. leaf 220
All fluorescent bacteria are construted by dear Rudolf (2018)
Reference
Schlechter, Rudolf O., Hyunwoo Jun, Michał Bernach, Simisola Oso, Erica Boyd, Dian A. Muñoz-Lintz, Renwick C. J. Dobson, Daniela M. Remus, and Mitja N. P. Remus-Emsermann. 2018. “Chromatic Bacteria - A Broad Host-Range Plasmid and Chromosomal Insertion Toolbox for Fluorescent Protein Expression in Bacteria.” Frontiers in Microbiology 9 (December): 3052.
Vinatzer, B. A., Teitzel, G. M., Lee, M.-W., Jelenska, J., Hotton, S., Fairfax, K., Jenrette, J., & Greenberg, J. T. (2006). The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Molecular Microbiology, 62(1), 26–44.